Fluorescent In Situ Hybridization (FISH)
FISH is the detection of highly specific DNA probes which have been hybridized to either interphase or metaphase chromosomes using fluorescence microscopy.
DNA for probe use is labeled with fluorescent molecules. The probes bind to a specific region or regions on the target chromosome. The chromosomes are then stained using a contrasting color, and the cells are viewed using a fluorescence microscope.
Different types of probes
Deletion
Unique sequence probes (sometimes referred to as LSI or “Locus-Specific Identifier probes) bind to specific small segments of certain chromosomes. These probes are useful in detection of microdeletion and microduplication syndromes.
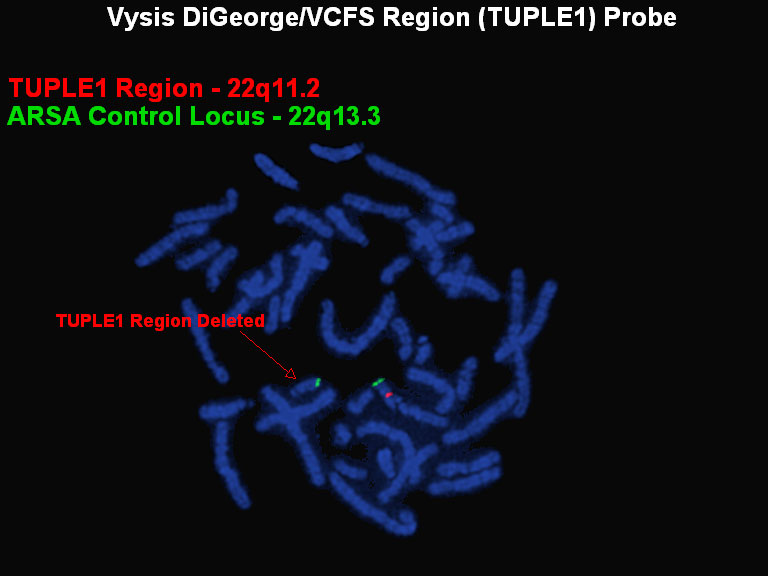
The red probe (TUPLE1) is specific to the DiGeorge/VCFS region just below the centromere of chromosome 22; the green probe is used as a control to identify the chromosome 22s. The chromosome on the right shows both the control and the DiGeorge/VCFS specific probe, the chromosome on the left shows only the control probe which indicates a deletion of the region to which the red probe would normally hybridize.
Duplication
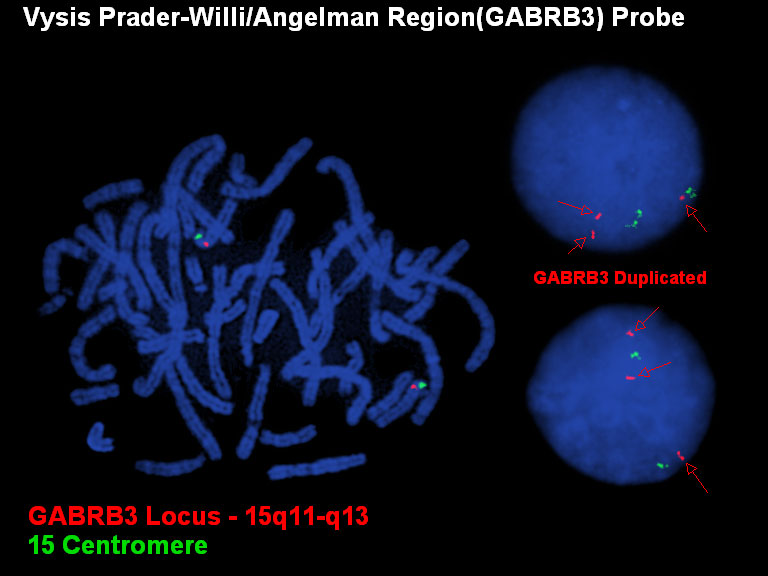
In this case there is a duplication rather than a deletion. There are three signals for the red probe (GABRB3 locus in 15q) and only two signals for the green control. This assay is done on interphase cells as the duplication is not always evident on metaphase chromosomes.
Fusion
Fusion probes are used to detect common translocations involving gene regions which, when rearranged and joined, result in leukemia and/or cancer.
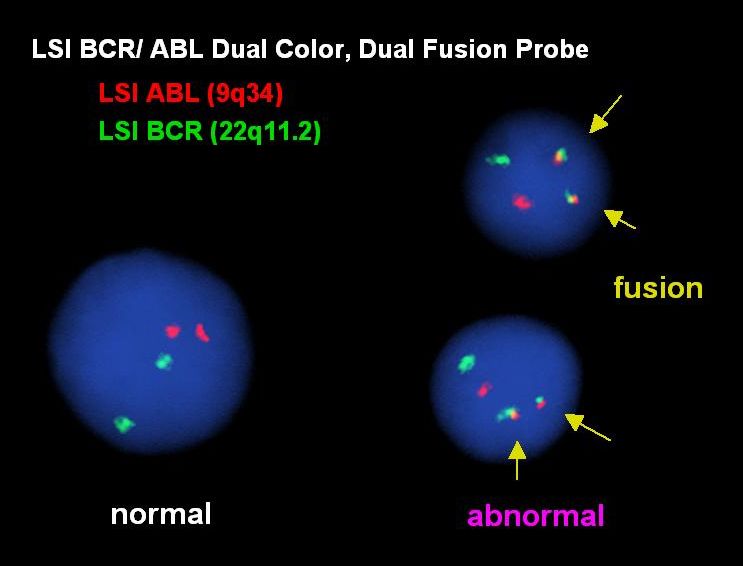
Dual fusion probes are used to detect specific translocations associated with cancer. The two genes involved in the translocation are labeled in different colors (ex: the ABL gene on chromosome 9 is red and the BCR gene on chromosome 22 is green), a nearby gene on each chromosome is also labeled in the same color. In normal cells we see two signals for each color (cell on the left). When the t(9;22)(q34;q11.2) [associated with CML] occurs one chromosome 9 and one chromosome 22 break, this separates the ABL gene from its nearby gene and separates the BCR gene from its nearby gene. The piece of chromosome 9 moves (translocates) to the chromosome 22, the piece of chromosome 22 moves to the chromosome 9; one chromosome has a fusion of the ABL and BCR genes, the other chromosome has a fusion of the two nearby genes (two red-green fused signals, cells on the right).
Break Apart
Some genes are involved in many different translocations associated with cancer, so instead of using a specific dual fusion probe to test for each possible translocation, a break apart probe is used to test whether the gene of interest is involved in a rearrangement. One end of the gene is labeled red and the other end of the gene is labeled green. Normal copies of the gene are seen as a red-green fused signal (cell on the right). If there is a translocation or rearrangement, the gene breaks into separate red and green signals.
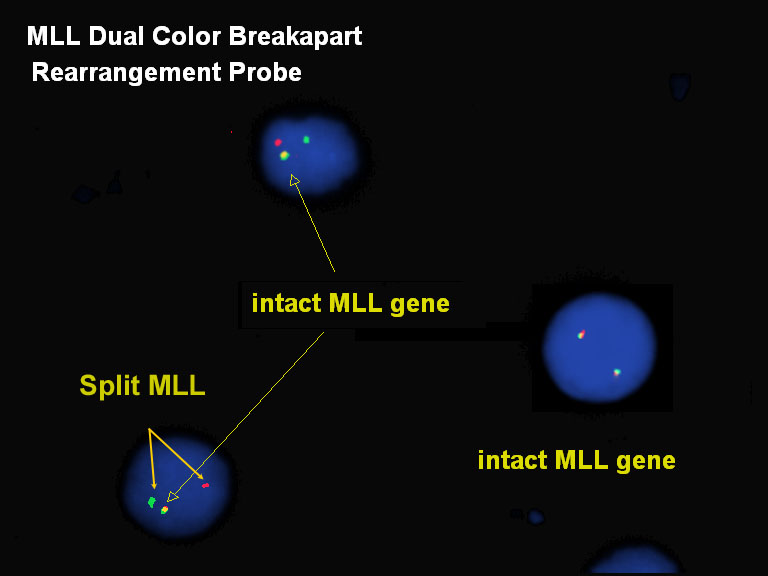
Here, the MLL gene on 11q23 is rearranged (but the translocation partner cannot be determined by this method alone).
Enumeration
Some cancers are associated with amplification of a gene; enumeration probes are used to detect the relative copy number.
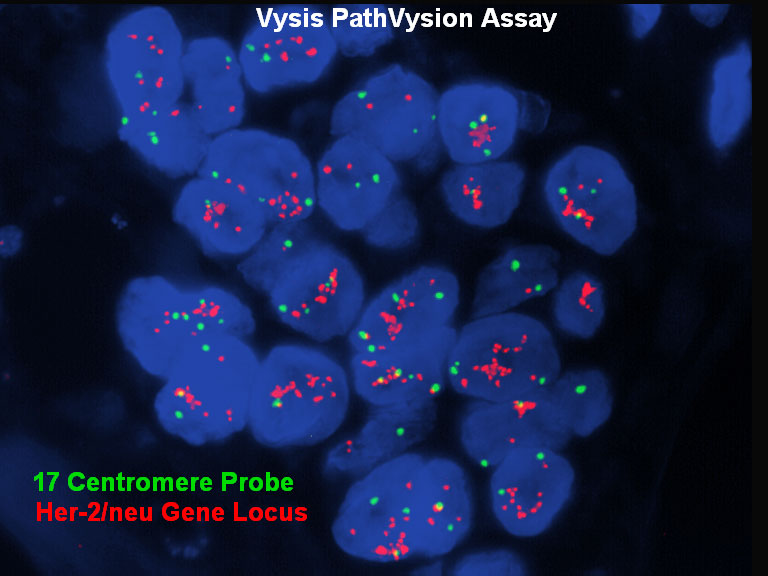
In some types of breast cancer, the cells acquire extra copies of the HER2 gene on chromosome 17q11.2-q12. For this test the HER2 gene is labeled red and the centromere of chromosome 17 is labeled green. In a normal cell there will be two signals for each color and a ratio of 1.0 (HER2:centromere). In this case the HER2 gene is amplified with many more red signals than green signals.
FISH Probes by Chromosome | FISH Probes by Usage | Back to Top of Page